What is Molecular Docking?
Molecular docking is a computational technique that uses various algorithms to predict the binding mode of a small molecule ligand to a receptor, typically a protein. The ligand can be any molecule that is known to bind to the protein, such as a drug candidate, while the receptor is a macromolecule, such as a protein or DNA. The process involves finding the most energetically favorable orientation of the two molecules to form a stable complex.
Why Do We Do Protein Docking?
Protein docking is an important tool in the drug discovery process, as it allows us to predict the binding of small molecule drugs to their target proteins. This is a crucial step in the development of new drugs, as it provides insight into the effectiveness of the drug candidate before any experimental testing is performed. Protein docking can also be used to understand the binding mechanism of a known drug, which can lead to the discovery of new drug targets and the development of more effective treatments for various diseases.

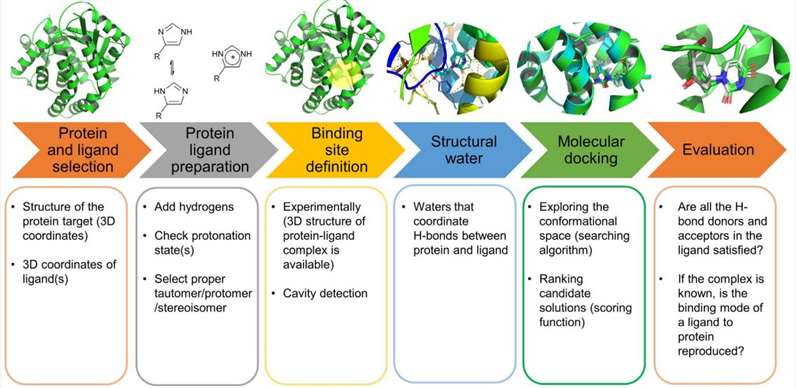
A typical docking workflow (Stanzione et al., 2021).
How Does Molecular Docking Work?
Molecular docking involves several steps, which can be broadly classified into two main categories: sampling and scoring.
- Sampling
Sampling involves generating different conformations of the ligand and receptor molecules and predicting their interactions in 3D space. Several algorithms can be used for this purpose, such as Monte Carlo and genetic algorithms.
- Scoring
Scoring involves evaluating the energetics of the predicted complexes and ranking them based on their binding affinity. The scoring function takes into account various factors such as van der Waals forces, electrostatic interactions, and hydrogen bonding.
 Molecular docking analysis of XAN (Acharya et al., 2019)
Molecular docking analysis of XAN (Acharya et al., 2019)
What is the Success Rate of Molecular Docking?
The success rate of molecular docking varies depending on several factors such as the size and complexity of the ligand and receptor molecules, the accuracy of the scoring function, and the sampling method used. In general, molecular docking has a success rate of around 30-40%, which means that only a small percentage of predicted complexes bind in the experimental setting.
What are the Benefits of Molecular Docking?
Molecular docking has several benefits, including:
- Cost-effective: Molecular docking is a cost-effective method for identifying potential drug candidates, as it eliminates the need for expensive and time-consuming experimental testing.
- High-throughput: Molecular docking can be used to screen large databases of compounds, allowing for the rapid identification of potential drug candidates.
- Insight into Binding Mechanisms: Molecular docking provides insight into the binding mechanism of a drug candidate, allowing for the development of more effective treatments for various diseases.
What is an Example of a Molecular Docking?
An excellent demonstration of the practical use of molecular docking is the progress in the development of HIV protease inhibitors. Specifically, HIV protease represents a pivotal enzyme in the replication of the HIV virus, and it constitutes a crucial drug target. In the late 1980s, researchers were able to exploit molecular docking to pinpoint compounds that would bind to the active site of the HIV protease enzyme, in a bid to create new drugs. Through this approach, they managed to develop powerful HIV/AIDS therapies, such as saquinavir and ritonavir, which have been critical in treating HIV/AIDS patients.
In a more recent example, researchers adopted a similar strategy, applying molecular docking to identify potential drug candidates for the treatment of COVID-19. Focusing on the SARS-CoV-2 main protease, which represents a pivotal enzyme in the replication of the virus, they leveraged molecular docking to screen a vast database of compounds in search of promising candidates. Thanks to this approach, they were able to pinpoint several exciting drug candidates, among them the FDA-approved drug lopinavir. Notably, this strategy provides a quicker and more cost-effective approach to identifying potential drug candidates for the treatment of COVID-19, a formidable challenge currently facing the medical community.
What To Do After Molecular Docking?
Following molecular docking, the subsequent phase in the drug discovery process typically entails experimentally validating the forecasted binding of the small molecule drug to the target protein. A diverse range of techniques such as X-ray crystallography, NMR spectroscopy, and biochemical assays can facilitate this validation process. These methods can aid in determining the precise binding site of the drug, the strength of the binding, as well as any possible side effects or toxicity issues.
Once the experimental verification of the drug’s binding has been carried out, the drug candidate can progress to further testing, such as pharmacokinetics and toxicity studies. This phase allows for the evaluation of drug metabolism, absorption, and excretion in the body, in addition to the investigation of any toxicological risks the drug may pose. The drug candidate must successfully pass these tests before advancing to clinical trials. Clinical trials, on the other hand, are designed to investigate the safety and efficacy of the drug in humans. However, it’s essential to note that clinical trials are an extensive and costly process.
References
- Stanzione, Francesca, Ilenia Giangreco, and Jason C. Cole. “Use of molecular docking computational tools in drug discovery.” Progress in Medicinal Chemistry 60 (2021): 273-343.
- Acharya, Reetuparna, et al. “Structure based multitargeted molecular docking analysis of selected furanocoumarins against breast cancer.” Scientific reports 9.1 (2019): 1-13.
Read More: Protein-Protein Interaction Analysis